This topic takes on average 55 minutes to read.
There are a number of interactive features in this resource:
 Chemistry
Chemistry
Getting ill usually means suffering from something fairly familiar – tonsillitis, a cold, ‘flu or pneumonia. But on the 17th of March 2003 the arrival of a new and potentially deadly disease was announced in the world media. SARS (severe acute respiratory syndrome) originated in the Guandong area of southern China but soon spread to Hong Kong and from there to countries such as Canada and the USA.
The early symptoms of SARS are fairly vague. The sufferer has a fever of over 38oC, and may have a headache, general body aching and discomfort – symptoms which could be any one of many familiar diseases. Over the first few days a dry cough and some difficulty breathing may develop. In many patients these symptoms gradually improve and they recover, but for some, particularly older people and people who have damaged immune systems, the infection can be fatal. SARS seems to be spread by close contact between people through droplet infection – most people who have fallen ill have lived with or cared for someone who is infected, or they have travelled on the same plane, close together for several hours.
A vital step in understanding how to treat, cure or prevent a new disease is knowing what causes it. This used to take months or even years. But within a week of the first outbreak of SARS scientists had a good idea what was causing it, and within a month they had sequenced the RNA of the virus involved. This was all made possible by PCR.
Specimens of body fluids taken from SARS patients were made available to scientists and the genetic material amplified using PCR. The RNA could then be sequenced and identified – the genetic material from the affected person and from known micro-organisms eliminated, leaving the genetic material of the microbe causing SARS. It has turned out to be a new mutation of a coronavirus.
PCR is also very important in diagnosing SARS. The problem is that the symptoms are very easy to mistake for other diseases, yet SARS can be fatal. The World Health Organisation (WHO) worked very hard to contain the disease and prevent it from spreading around the world. This meant it was very important to know if someone is suffering from SARS so they and their contacts can be kept in isolation.
Diagnostic tests using antibodies are so far only effective about 10 days after infection – by which time the person will be either in intensive care, dead – or getting better. PCR enables doctors and scientists to get a much more rapid diagnosis. Two German scientists used PCR to amplify and so identify unique sections of the viral genetic material. They have developed probes which will attach to these sections and show them up. So material from a patient with suspected SARS is processed using PCR, and then the amplified genetic material is exposed to the SARS coronavirus specific probes. This can show up infection before the disease has progressed too far, making it easier to provide effective treatment and to avoid further cross infection.
Dr Klaus Stuhr of the WHO emphasised the way in which scientists throughout the international community worked together, sharing results and ideas, to enable such rapid progress in identifying the SARS virus to be made. All the usual competition to be the first to publish results was set aside in the face of this global health emergency. The great fear was that SARS might spread to the African nations, where so many people already have weakened immune systems from the effects of malnutrition, malaria and AIDS. It could wipe out millions of people in a very short time.
The tests are still being refined and made more accurate – they are not completely reliable and there are even ideas that more than one virus might be involved. But thanks in no small part to the polymerase chain reaction, doctors and scientists managed to control this disease and prevent it becoming a major disaster – the last recorded outbreak by the WHO was in China in 2004.
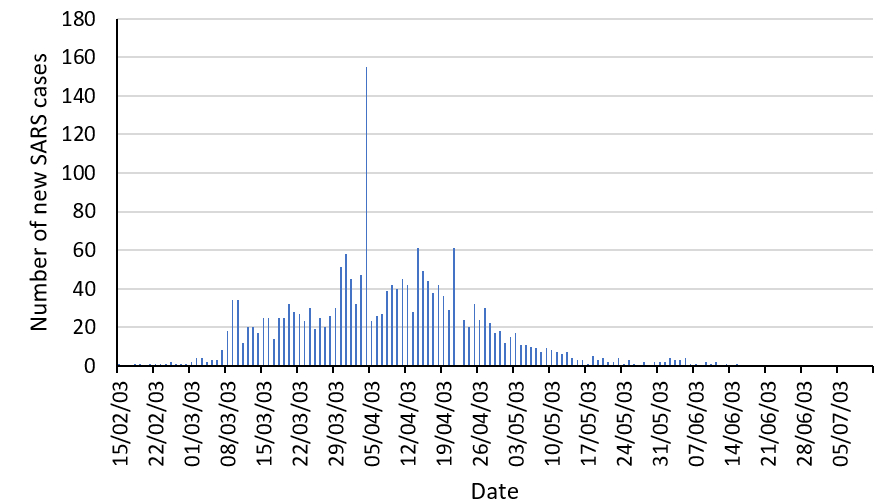
On February 21st 2003 a doctor from Guangdong Province in China flew to Hong Kong and stayed one night in a hotel room, visiting his family. He was unwell when he set off from China, and by the 22nd of February he was seriously ill and admitted to hospital in Hong Kong, where he died of SARS 10 days later. In that short time, 12 other guests in the hotel became infected. Two of them were admitted to local hospitals, triggering local outbreaks of the disease. As a result of easy international travel, other guests and hotel visitors seeded outbreaks of SARS in countries as far apart as Vietnam, Cambodia and Canada, when they continued their journeys.
The Hong Kong health authorities implemented strict infection control policies, identifying the pathogen and isolating infected patients from the population at large, and the outbreak peaked towards the end of March. By the end of June, the outbreak was over and the WHO removed Hong Kong from its list of areas infected by SARS. There had been 1755 cases, and 295 people had died – a mortality rate of 16.8%. Around 30% of the people affected were healthcare workers.
Hong Kong was a microcosm of SARS globally, demonstrating how easily the virus spread and the role of international travel in the spread of disease from a single infected individual. It was also a model of how to contain a new infection, demonstrating how identification of the pathogen and isolation of infected individuals successfully eradicated the disease.
Tuberculosis is a very ancient disease – we have clear evidence, using PCR and genome sequencing that Egyptian mummies died of tuberculosis in 600 BC. Caused by the slow-growing Mycobacterium tuberculosis and M.bovis (from cattle), globally almost nine million people are affected by TB, and over a million die each year. TB is not easy to diagnose. The bacteria which cause it are very hard to culture, and even when they grow successfully, it is a very slow process and it can take weeks before the bacteria can be positively identified. During this time, people can still spread the disease. Another problem is that the rate of infection with antibiotic-resistant strains of the bacteria has increased steadily. Doctors prescribe by trial and error until they find the most effective antibiotic – a process which not only prolongs disease and increases the spread of the bacteria, but also increases the risk of further antibiotic resistance developing.
In 2017, British scientists made a breakthrough. Using a combination of real time-PCR and the latest genome sequencing technologies, they developed a diagnostic test which gives them the specific type of bacterium in a suspected TB case within days of collecting the sample and this also tells them the antibiotics to which it is vulnerable. Using this technique doctors should be able to diagnose the disease rapidly and accurately. This means the people who need treatment for TB will get it fast and they will be treated with antibiotics which work. In turn this will cure the patients faster, reduce the spread of disease and minimise the development of further antibiotic resistance. The cost and speed of the diagnostic test are also set to reduce rapidly over the next few years – it’s a win-win-win situation in the battle against M tuberculosis - one of our oldest adversaries.
Ebola is a rare and deadly disease caused by the Ebola virus. The first recorded outbreak was in 1976, in Zaire, also known as the Democratic Republic of the Congo – and 88% of the people infected died. So far, five Ebola species have been discovered. They are all animal viruses, and four of them can be transmitted to humans and cause human disease. People become infected through direct contact (through broken skin, or mucous membranes in the mouth, nose, eyes and genitals) with any of the following:
blood or body fluids (eg blood, faeces, saliva, sweat, vomit, breast milk, semen) from someone who is infected with Ebola or has died of Ebola
infected fruit bats or apes or monkeys (primates) who are infected with Ebola (through bites, blood, meat, faeces or urine)
needles or syringes contaminated with body fluids from someone infected with Ebola or who has died from Ebola
scientists are investigating whether it is possible to become infected with Ebola through the semen of someone who has recovered from Ebola.
The most recent outbreak of Ebola started in West Africa in late 2013, although it was 2014 before it was recognised. It spread to many countries including Liberia, Sierra Leone, Nigeria and Guinea, with people affected as far away as the UK, Spain and Italy. The WHO finally declared the outbreak over in March 2016. PCR played a direct role in helping identify people who were infected, and in helping to develop vaccines against the deadly disease. Scientists are also using PCR to develop testing kits which can be used out in the field to help identify people infected by Ebola as soon as possible. A cheap, effective test would enable health professionals to isolate people quickly, reducing the risk of the virus spreading.

The story of Ebola and the battle of scientists to overcome this threat to human health is still continuing. Use the resources here and elsewhere on the Web (for example on the CDC Ebola page) to help you find answers to the questions below. In each case, consider the role that PCR has played or might play in future.
How many people were affected by Ebola in the 2014-16 out-break and how many died from the disease?
How did the diagnostic tests for Ebola work, and why are effective diagnostic tests so important in helping to prevent outbreaks of Ebola in the future?
How close are scientists to developing an effective vaccine against Ebola?
Resources for research into Ebola are limited. Discuss the pros and cons of using this limited funding for
Students may use the above resources and present their findings in an open discussion or debate format, or in the shape of a news report for a non-scientific audience or a scientific article.
