This topic takes on average 55 minutes to read.
There are a number of interactive features in this resource:
 Biology
Biology
 PSHE / Citizenship studies
PSHE / Citizenship studies
 Science (applied)
Science (applied)
In the early 1970s, the first genetically engineered organism was a bacterium - E. coli. The techniques used for genetic engineering are changing all the time. The basic principles of classic genetic engineering are shown here. The recently developed CRISPR-Cas9 gene editing process is covered later.
Stage 1: The desired gene can be removed from the DNA of the donor organism using enzymes called restriction endonucleases. These are enzymes which chop up DNA strands by cutting them at specific sites, so they can be used to remove very specific genes. Certain types of restriction endonucleases are particularly useful because they leave small regions of DNA sticking out at each end of the required gene. These are known as sticky ends, and make it much easier to attach the gene into another piece of DNA.
Sometimes the required gene is synthesised artificially. Using another specialised enzyme known as reverse transcriptase, the DNA sequence of the gene can be built up from isolated pieces of mRNA which have already been transcribed from the required gene.

Stage 2: The second step is to prepare a vector molecule to carry the DNA into the host cell – often a bacterium. A bacterial plasmid (a small circular strand of DNA often found in bacteria in addition to their main DNA) is often used as a vector. These replicate quickly and independently of the main bacterial genome, and can therefore amplify the number of copies of the gene.
Plasmids usually carry a marker gene which is used to demonstrate the cells which have been successfully engineered. In the early days, these marker genes often coded for characteristics such as resistance to a particular antibiotic. However, there were many concerns about the use of antibiotic resistance as marker genes, including the risk of them crossing into pathogens. In response, scientists modified their techniques and now other characteristics, such as fluorescence or the ability to synthesise a specific nutrient are used to identify the genetically modified organisms. So, for example, bacteria can be grown in a medium with a particular nutrient missing to show which cells have been successfully engineered: only the genetically-engineered organisms will be able to synthesise the missing nutrient and so only they will grow.
The bacterial plasmid is opened up using restriction enzymes which leave sticky ends that correspond to those of the new gene.
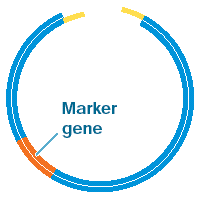
Stage 3: The third step is to join the new gene into the bacterial plasmid. The sticky ends are lined up and the gene is attached or annealed – using enzymes called DNA ligases which join the pieces of DNA together.
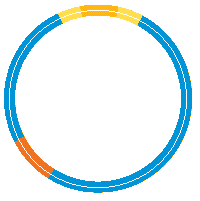
Stage 4: The final step is to incorporate the engineered DNA into the bacterium or other cell where it is required. This is known as transformation and is usually achieved by suddenly heating up the bacteria which makes their cell membrane more permeable so plasmid can move into the cells. Once the plasmid is inside the host bacterium it will be expressed and a new protein made.

Microorganisms are the most commonly used organisms in genetic engineering because they are relatively easy, quick and cheap to culture and there are few ethical issues about their usage. This entire process can either be used to generate a genetically engineered product, or as just a step of the genetic engineering process to clone the DNA (make many copies of it) before it is studied in more detail and introduced into a more complex organism.
To change the DNA of eukaryotic organisms such as people, a different vector must be used (usually a virus as human cells don’t contain plasmids), the correct cells must be targeted, the gene must be activated once it is inside the cell, the gene must be integrated correctly, and all these things must occur without harmful side effects. Engineering eukaryotic cells is often more complex than modifying bacteria, but as techniques develop it is becoming increasingly common.